Difference between revisions of "DiversityDescriptions Quiz (Artenquiz)"
| (12 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | The ER diagram of DiversityDescriptions_Quiz (DD_Quiz) is a clone of [[DescriptionsModel v3.00.10|DiversityDescriptions 3.00.10. July 17th, 2014]] that is intended to work with the [http://biowikifarm.net/meta/Django-Installation Web-Framework Django] to have a quiz for species descriptions in German. This project is presently under development. Additional tables see below. Suggested citation is that of DiversityDescriptions 3.00.10: G. Hagedorn, A. Plank, A. Link, G. Rambold & D. Triebel (2014). DiversityDescriptions information model (version 3.00.10). http://www.diversityworkbench.net/Portal/DescriptionsModel_v3.00.10 | + | The ER diagram of DiversityDescriptions_Quiz (DD_Quiz) is a clone of [[DescriptionsModel v3.00.10|DiversityDescriptions 3.00.10. July 17th, 2014]] that is intended to work with the [http://biowikifarm.net/meta/Django-Installation Web-Framework Django] to have a quiz for species descriptions in German. This project is presently under development. Additional tables see below. For further information see under [[DiversityDescriptions Implementations|DiversityDescriptions Quiz implementations]]. |
| + | |||
| + | |||
| + | Suggested citation is that of DiversityDescriptions 3.00.10: G. Hagedorn, A. Plank, A. Link, G. Rambold & D. Triebel (2014). DiversityDescriptions information model (version 3.00.10). http://www.diversityworkbench.net/Portal/DescriptionsModel_v3.00.10. | ||
| Line 6: | Line 9: | ||
== Documentation == | == Documentation == | ||
=== Database === | === Database === | ||
| + | |||
| + | Scheme of DiversityDescriptions Quiz: | ||
<graphviz caption='Legend of DiversityDescriptions datebase scheme' alt='Legend of DiversityDescriptions' format='png'> | <graphviz caption='Legend of DiversityDescriptions datebase scheme' alt='Legend of DiversityDescriptions' format='png'> | ||
| Line 30: | Line 35: | ||
The following conventions and abbreviations have been used in the tables: Columns of primary key:<u>underlined</u>. Categorical data are presented to a user in the quiz therefore some tables (in <span style="color:gray">gray</span>) are not used for the quiz. | The following conventions and abbreviations have been used in the tables: Columns of primary key:<u>underlined</u>. Categorical data are presented to a user in the quiz therefore some tables (in <span style="color:gray">gray</span>) are not used for the quiz. | ||
| + | [[File:DiversityDescriptions Quiz Artenquiz digraph DD Quiz Scheme 20140717.png|thumb|center|1000px|Database Scheme of DiversityDescriptions Quiz Artenquiz (20140717)]] | ||
| + | <div style="clear:both"></div> | ||
| + | |||
| + | <div class="mw-collapsible mw-collapsed"> | ||
| + | The data base scheme described in DOT language (click the expand on the right side to read the content) | ||
| + | <pre class="mw-collapsible-content"> | ||
<graphviz caption='Scheme of Diversity Descriptions Quiz' alt='Diversity Descriptions Quiz' format='png'> | <graphviz caption='Scheme of Diversity Descriptions Quiz' alt='Diversity Descriptions Quiz' format='png'> | ||
| Line 38: | Line 49: | ||
Annotation [ | Annotation [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Annotation" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Annotation]]" |
tooltip="One record per annotated object (Char, State, Descr. etc.)" | tooltip="One record per annotated object (Char, State, Descr. etc.)" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 55: | Line 66: | ||
]; | ]; | ||
BaseEntity [ | BaseEntity [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_BaseEntity" | + | URL="[[DescriptionsModel_v3.00.10#Table:_BaseEntity]]" |
tooltip="The BaseEntity is used within the database to provide unique keys" | tooltip="The BaseEntity is used within the database to provide unique keys" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0"> | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0"> | ||
| Line 67: | Line 78: | ||
]; | ]; | ||
BaseEntityTable_Enum [ | BaseEntityTable_Enum [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_BaseEntityTable_Enum" | + | URL="[[DescriptionsModel_v3.00.10#Table:_BaseEntityTable_Enum]]" |
tooltip="The BaseEntityTable_Enum contains the names of tables that reference the BaseEntity table" | tooltip="The BaseEntityTable_Enum contains the names of tables that reference the BaseEntity table" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 78: | Line 89: | ||
]; | ]; | ||
CategoricalSamplingData [ | CategoricalSamplingData [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_CategoricalSamplingData" | + | URL="[[DescriptionsModel_v3.00.10#Table:_CategoricalSamplingData]]" |
tooltip="The categorical data recorded for a sampling event" | tooltip="The categorical data recorded for a sampling event" | ||
fontcolor="gray" | fontcolor="gray" | ||
| Line 94: | Line 105: | ||
]; | ]; | ||
CategoricalState [ | CategoricalState [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_CategoricalState" | + | URL="[[DescriptionsModel_v3.00.10#Table:_CategoricalState]]" |
tooltip="The categorical states available for categorical descriptors" | tooltip="The categorical states available for categorical descriptors" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 110: | Line 121: | ||
]; | ]; | ||
CategoricalSummaryData [ | CategoricalSummaryData [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_CategoricalSummaryData" | + | URL="[[DescriptionsModel_v3.00.10#Table:_CategoricalSummaryData]]" |
tooltip="The categorical data of a description" | tooltip="The categorical data of a description" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 125: | Line 136: | ||
]; | ]; | ||
Contribution [ | Contribution [ | ||
| − | URL="#Table:_Contribution" | + | URL="[[#Table:_Contribution]]" |
tooltip="Multiple contributors may contribute in different roles" | tooltip="Multiple contributors may contribute in different roles" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 138: | Line 149: | ||
]; | ]; | ||
Contributor [ | Contributor [ | ||
| − | URL="#Table:_Contributor" | + | URL="[[#Table:_Contributor]]" |
tooltip="People contibuting to a work. Roles are recorded in Contribution" | tooltip="People contibuting to a work. Roles are recorded in Contribution" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 153: | Line 164: | ||
]; | ]; | ||
DataStatus_Enum [ | DataStatus_Enum [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DataStatus_Enum" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DataStatus_Enum]]" |
tooltip="Values of data status used for descriptions according to SDD 1.1 rev 5" | tooltip="Values of data status used for descriptions according to SDD 1.1 rev 5" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 169: | Line 180: | ||
]; | ]; | ||
Description [ | Description [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Description" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Description]]" |
tooltip="The description in the database" | tooltip="The description in the database" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 181: | Line 192: | ||
]; | ]; | ||
DescriptionScope [ | DescriptionScope [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DescriptionScope" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DescriptionScope]]" |
tooltip="The scope of the description" | tooltip="The scope of the description" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 200: | Line 211: | ||
]; | ]; | ||
Descriptor [ | Descriptor [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Descriptor" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Descriptor]]" |
tooltip="Descriptor (= characters, features) define variables" | tooltip="Descriptor (= characters, features) define variables" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 227: | Line 238: | ||
]; | ]; | ||
DescriptorInapplicability [ | DescriptorInapplicability [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DescriptorInapplicability" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorInapplicability]]" |
tooltip="The descriptor dependency rules" | tooltip="The descriptor dependency rules" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 240: | Line 251: | ||
]; | ]; | ||
DescriptorMapping_C2C [ | DescriptorMapping_C2C [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DescriptorMapping_C2C" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorMapping_C2C]]" |
tooltip="Mapping of a categorical state to another categorical state" | tooltip="Mapping of a categorical state to another categorical state" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 251: | Line 262: | ||
]; | ]; | ||
DescriptorMapping_Q2C [ | DescriptorMapping_Q2C [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DescriptorMapping_Q2C" fontcolor="gray" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorMapping_Q2C]]" fontcolor="gray" |
tooltip="Mapping of a quantitative descriptor to a categorical state" | tooltip="Mapping of a quantitative descriptor to a categorical state" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | ||
| Line 266: | Line 277: | ||
]; | ]; | ||
DescriptorStatusData [ | DescriptorStatusData [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DescriptorStatusData" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorStatusData]]" |
tooltip="The status data of a descriptor for a certain description" | tooltip="The status data of a descriptor for a certain description" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 280: | Line 291: | ||
]; | ]; | ||
DescriptorTree [ | DescriptorTree [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DescriptorTree" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorTree]]" |
tooltip="The root and definition of a descriptor tree" | tooltip="The root and definition of a descriptor tree" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 294: | Line 305: | ||
]; | ]; | ||
DescriptorTreeNode [ | DescriptorTreeNode [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DescriptorTreeNode" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorTreeNode]]" |
tooltip="The descriptor tree nodes representing either nodes of the tree or descriptors ("leafes" of the tree)" | tooltip="The descriptor tree nodes representing either nodes of the tree or descriptors ("leafes" of the tree)" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 310: | Line 321: | ||
]; | ]; | ||
DescriptorTreeNodeRecFrequency [ | DescriptorTreeNodeRecFrequency [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DescriptorTreeNodeRecFrequency" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorTreeNodeRecFrequency]]" |
tooltip="Selection of recommended frequency values for descriptor tree parts or single descriptors" | tooltip="Selection of recommended frequency values for descriptor tree parts or single descriptors" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 322: | Line 333: | ||
]; | ]; | ||
DescriptorTreeNodeRecModifier [ | DescriptorTreeNodeRecModifier [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DescriptorTreeNodeRecModifier" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorTreeNodeRecModifier]]" |
tooltip="Selection of recommended modifier values for descriptor tree parts or single descriptors" | tooltip="Selection of recommended modifier values for descriptor tree parts or single descriptors" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 334: | Line 345: | ||
]; | ]; | ||
DescriptorTreeNodeRecStatMeasure [ | DescriptorTreeNodeRecStatMeasure [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_DescriptorTreeNodeRecStatMeasure" | + | URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorTreeNodeRecStatMeasure]]" |
tooltip="Selection of recommended statistical measures for descriptor tree parts or single descriptors" | tooltip="Selection of recommended statistical measures for descriptor tree parts or single descriptors" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 346: | Line 357: | ||
]; | ]; | ||
Frequency [ | Frequency [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Frequency" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Frequency]]" |
tooltip="Definition of frequency modifier values" | tooltip="Definition of frequency modifier values" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 360: | Line 371: | ||
]; | ]; | ||
GeographicAreaProxy [ | GeographicAreaProxy [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_GeographicAreaProxy" | + | URL="[[DescriptionsModel_v3.00.10#Table:_GeographicAreaProxy]]" |
tooltip="Geographic Area, proxy for WB component" | tooltip="Geographic Area, proxy for WB component" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 371: | Line 382: | ||
]; | ]; | ||
GeographicArea_Taxon [ | GeographicArea_Taxon [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_GeographicArea_Taxon" | + | URL="[[DescriptionsModel_v3.00.10#Table:_GeographicArea_Taxon]]" |
tooltip="Presence (or likely presence) of taxon in a given geographic area" | tooltip="Presence (or likely presence) of taxon in a given geographic area" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 383: | Line 394: | ||
]; | ]; | ||
Identifier [ | Identifier [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Identifier" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Identifier]]" |
tooltip="Objects may have multiple external identifiers" | tooltip="Objects may have multiple external identifiers" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 394: | Line 405: | ||
]; | ]; | ||
Modifier [ | Modifier [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Modifier" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Modifier]]" |
tooltip="Definition of modifier values" | tooltip="Definition of modifier values" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 407: | Line 418: | ||
]; | ]; | ||
ObservationProxy [ | ObservationProxy [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_ObservationProxy" | + | URL="[[DescriptionsModel_v3.00.10#Table:_ObservationProxy]]" |
tooltip="Observation; proxy for WB component" | tooltip="Observation; proxy for WB component" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 418: | Line 429: | ||
]; | ]; | ||
OrganismRelationData [ | OrganismRelationData [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_OrganismRelationData" | + | URL="[[DescriptionsModel_v3.00.10#Table:_OrganismRelationData]]" |
tooltip="Categorical data (states recorded for a description)" | tooltip="Categorical data (states recorded for a description)" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 433: | Line 444: | ||
]; | ]; | ||
OtherScope [ | OtherScope [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_OtherScope" | + | URL="[[DescriptionsModel_v3.00.10#Table:_OtherScope]]" |
tooltip="Scope values for description scope values for scope types 'Other scope', 'Part', 'Stage' and 'Sex' " | tooltip="Scope values for description scope values for scope types 'Other scope', 'Part', 'Stage' and 'Sex' " | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 447: | Line 458: | ||
]; | ]; | ||
Project [ | Project [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Project" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Project]]" |
tooltip="Projects define separated workareas in a single database" | tooltip="Projects define separated workareas in a single database" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 466: | Line 477: | ||
]; | ]; | ||
Project_AvailableScope [ | Project_AvailableScope [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Project_AvailableScope" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Project_AvailableScope]]" |
tooltip="Scope values available for a certain project" | tooltip="Scope values available for a certain project" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 478: | Line 489: | ||
]; | ]; | ||
QuantitativeSamplingData [ | QuantitativeSamplingData [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_QuantitativeSamplingData" | + | URL="[[DescriptionsModel_v3.00.10#Table:_QuantitativeSamplingData]]" |
tooltip="The quantitative data recorded for a sampling event" | tooltip="The quantitative data recorded for a sampling event" | ||
fontcolor="gray" | fontcolor="gray" | ||
| Line 494: | Line 505: | ||
]; | ]; | ||
QuantitativeSummaryData [ | QuantitativeSummaryData [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_QuantitativeSummaryData" | + | URL="[[DescriptionsModel_v3.00.10#Table:_QuantitativeSummaryData]]" |
tooltip="The quantitative data recorded for a sampling event" fontcolor="gray" | tooltip="The quantitative data recorded for a sampling event" fontcolor="gray" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | ||
| Line 510: | Line 521: | ||
]; | ]; | ||
ReferenceProxy [ | ReferenceProxy [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_ReferenceProxy" | + | URL="[[DescriptionsModel_v3.00.10#Table:_ReferenceProxy]]" |
tooltip="Reference (literature/source); proxy for WB component" | tooltip="Reference (literature/source); proxy for WB component" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 521: | Line 532: | ||
]; | ]; | ||
Resource [ | Resource [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Resource" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Resource]]" |
tooltip="Hyperlinks to separate rich text/media objects" | tooltip="Hyperlinks to separate rich text/media objects" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 543: | Line 554: | ||
]; | ]; | ||
ResourceVariant [ | ResourceVariant [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_ResourceVariant" | + | URL="[[DescriptionsModel_v3.00.10#Table:_ResourceVariant]]" |
tooltip="Different resource variants/instances/service access points" | tooltip="Different resource variants/instances/service access points" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 561: | Line 572: | ||
]; | ]; | ||
ResourceVariant_Enum [ | ResourceVariant_Enum [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_ResourceVariantEnum" | + | URL="[[DescriptionsModel_v3.00.10#Table:_ResourceVariantEnum]]" |
tooltip="Classes for resource variants, values are predefined in the database" | tooltip="Classes for resource variants, values are predefined in the database" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 577: | Line 588: | ||
ResourceWatchedTime [ | ResourceWatchedTime [ | ||
| − | URL="#Table:_ResourceWatchedTime" | + | URL="[[#Table:_ResourceWatchedTime]]" |
tooltip="The time a resource thumb was watched (=mouse hover)" | tooltip="The time a resource thumb was watched (=mouse hover)" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 589: | Line 600: | ||
SamplingEvent [ | SamplingEvent [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_SamplingEvent" | + | URL="[[DescriptionsModel_v3.00.10#Table:_SamplingEvent]]" |
tooltip="A sampling event may contain many sampling units" fontcolor="gray" | tooltip="A sampling event may contain many sampling units" fontcolor="gray" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | ||
| Line 608: | Line 619: | ||
]; | ]; | ||
SamplingUnit [ | SamplingUnit [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_SamplingUnit" | + | URL="[[DescriptionsModel_v3.00.10#Table:_SamplingUnit]]" |
tooltip="Categorical data" fontcolor="gray" | tooltip="Categorical data" fontcolor="gray" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COlOR="gray"> | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COlOR="gray"> | ||
| Line 618: | Line 629: | ||
]; | ]; | ||
SexStatus_Enum [ | SexStatus_Enum [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_SexStatus_Enum" | + | URL="[[DescriptionsModel_v3.00.10#Table:_SexStatus_Enum]]" |
tooltip="Values of sex status predefined according to SDD 1.1 rev 5" | tooltip="Values of sex status predefined according to SDD 1.1 rev 5" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 634: | Line 645: | ||
]; | ]; | ||
SpecimenProxy [ | SpecimenProxy [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_SpecimenProxy" | + | URL="[[DescriptionsModel_v3.00.10#Table:_SpecimenProxy]]" |
tooltip="Specimens from collections; proxy for WB component" fontcolor="gray" | tooltip="Specimens from collections; proxy for WB component" fontcolor="gray" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | ||
| Line 645: | Line 656: | ||
]; | ]; | ||
StatisticalMeasure_Enum [ | StatisticalMeasure_Enum [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_StatisticalMeasure_Enum" fontcolor="gray" | + | URL="[[DescriptionsModel_v3.00.10#Table:_StatisticalMeasure_Enum]]" fontcolor="gray" |
tooltip="The statistical measures predefined according SDD 1.1 rev 5" | tooltip="The statistical measures predefined according SDD 1.1 rev 5" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | ||
| Line 660: | Line 671: | ||
]; | ]; | ||
Tag [ | Tag [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Tag" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Tag]]" |
tooltip="Objects may have multiple tags (= keywords)" | tooltip="Objects may have multiple tags (= keywords)" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 672: | Line 683: | ||
]; | ]; | ||
TaxonProxy [ | TaxonProxy [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_TaxonProxy" | + | URL="[[DescriptionsModel_v3.00.10#Table:_TaxonProxy]]" |
tooltip="Taxon/tax. hierarchy (simplified); proxy for WB component" | tooltip="Taxon/tax. hierarchy (simplified); proxy for WB component" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 706: | Line 717: | ||
]; | ]; | ||
TaxonRank_Enum [ | TaxonRank_Enum [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_TaxonRank_Enum" | + | URL="[[DescriptionsModel_v3.00.10#Table:_TaxonRank_Enum]]" |
tooltip="Ranks for taxa (species, genus, family, order, class, etc.)" | tooltip="Ranks for taxa (species, genus, family, order, class, etc.)" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 721: | Line 732: | ||
]; | ]; | ||
TaxonSet [ | TaxonSet [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_TaxonSet" | + | URL="[[DescriptionsModel_v3.00.10#Table:_TaxonSet]]" |
tooltip="Groups (or sets) of taxa; including non-phylogenetic groups " | tooltip="Groups (or sets) of taxa; including non-phylogenetic groups " | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 734: | Line 745: | ||
]; | ]; | ||
TaxonSet_Taxon [ | TaxonSet_Taxon [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_TaxonSet_Taxon" | + | URL="[[DescriptionsModel_v3.00.10#Table:_TaxonSet_Taxon]]" |
tooltip="Membership of taxa in a taxon set (n:m relation)" | tooltip="Membership of taxa in a taxon set (n:m relation)" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 746: | Line 757: | ||
]; | ]; | ||
TextDescriptorData [ | TextDescriptorData [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_TextDescriptorData" | + | URL="[[DescriptionsModel_v3.00.10#Table:_TextDescriptorData]]" |
tooltip="The text data of a description" | tooltip="The text data of a description" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 760: | Line 771: | ||
]; | ]; | ||
TextSamplingData [ | TextSamplingData [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_TextSamplingData" | + | URL="[[DescriptionsModel_v3.00.10#Table:_TextSamplingData]]" |
tooltip="Free-form text data recorded for a sampling event" fontcolor="gray" | tooltip="Free-form text data recorded for a sampling event" fontcolor="gray" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray"> | ||
| Line 783: | Line 794: | ||
]; | ]; | ||
Translation [ | Translation [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_Translation" | + | URL="[[DescriptionsModel_v3.00.10#Table:_Translation]]" |
tooltip="The translations of entries related to BaseEntity" | tooltip="The translations of entries related to BaseEntity" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 797: | Line 808: | ||
]; | ]; | ||
TranslationColumn_Enum [ | TranslationColumn_Enum [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_TranslationColumn_Enum" | + | URL="[[DescriptionsModel_v3.00.10#Table:_TranslationColumn_Enum]]" |
tooltip="The TranslationColumn_Enum contains the column names that are translated in the Translation table" | tooltip="The TranslationColumn_Enum contains the column names that are translated in the Translation table" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 808: | Line 819: | ||
]; | ]; | ||
ProjectProxy [ | ProjectProxy [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_ProjectProxy" | + | URL="[[DescriptionsModel_v3.00.10#Table:_ProjectProxy]]" |
tooltip="The projects as stored in the module DiversityProjects" | tooltip="The projects as stored in the module DiversityProjects" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 819: | Line 830: | ||
]; | ]; | ||
UserProxy [ | UserProxy [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_UserProxy" | + | URL="[[DescriptionsModel_v3.00.10#Table:_UserProxy]]" |
tooltip="The user as stored in the module DiversityAgents" | tooltip="The user as stored in the module DiversityAgents" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 831: | Line 842: | ||
]; | ]; | ||
ProjectUser [ | ProjectUser [ | ||
| − | URL="DescriptionsModel_v3.00.10#Table:_ProjectUser" | + | URL="[[DescriptionsModel_v3.00.10#Table:_ProjectUser]]" |
tooltip="The projects that a user can access" | tooltip="The projects that a user can access" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 843: | Line 854: | ||
TaxonQuiz [ | TaxonQuiz [ | ||
| − | URL="#Table:_TaxonQuiz" | + | URL="[[#Table:_TaxonQuiz]]" |
tooltip="User’s correctly answered taxa" | tooltip="User’s correctly answered taxa" | ||
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" > | ||
| Line 973: | Line 984: | ||
} | } | ||
</graphviz> | </graphviz> | ||
| + | </pre> | ||
| + | </div> | ||
| Line 1,060: | Line 1,073: | ||
== New projects == | == New projects == | ||
| + | |||
| + | Schematic important relations for a project: | ||
| + | Project | ||
| + | ├ code (used for URL) | ||
| + | ├ label (directly used for user display and in text context) | ||
| + | ├ TaxonSet (i.e. taxa, media-resources etc.) | ||
| + | ├ DescritorTree (i.e. characters and its states, media-resources etc.) | ||
| + | ├ … | ||
| + | … | ||
Steps to enter data for a new project: | Steps to enter data for a new project: | ||
| − | Create a project (table: Project) | + | # Create a project (table: Project) |
| − | * Project.code is important: it is used as URL part and within the Django application as internal project identifier it should never be changed | + | #* Project.code is important: it is used as URL part and within the Django application as internal project identifier it should never be changed |
| − | * eventually create or associate scopes, like “female”, “male” (table: OtherScope, Project_AvailableScope) | + | #* eventually create or associate scopes, like “female”, “male” (table: OtherScope, Project_AvailableScope) |
| − | Eventually create the taxa (table: TaxonProxy) | + | # Eventually create the taxa (table: TaxonProxy) |
| − | * associate taxa to the project (tables: TaxonSet, TaxonSet_Taxon) | + | #* associate taxa to the project (tables: TaxonSet, TaxonSet_Taxon) |
| − | * associate images of taxa (tables: Resource, ResourceVariant, BaseEntity.specific_rights_text should contain the corresponding picture’s author name like: “© Author Name”). A set of maximum of 16 thumbs will be displayed | + | #* associate images of taxa (tables: Resource, ResourceVariant, BaseEntity.specific_rights_text should contain the corresponding picture’s author name like: “© Author Name”). A set of maximum of 16 thumbs will be displayed |
| − | * | + | #*# an image is associated correctly, when 1 taxon has 1 Resource, but 2 (or multiple) ResourceVariant(!!) |
| − | * eventually associate geographic area (table: GeographicAreaProxy, GeographicArea_Taxon) | + | #*# resource variants as ReourceVariant_Enum.code "tiny_sample" (=thumb 75×75px) and "best_quality" (max 500×500px displayed) should have corresponding urls in ResourceVariant.url:<!-- |
| − | Create character and character states (table Descriptor, CategoricalState), character tree (tables: DescriptorTree, DescriptorTreeNode) | + | --><code><br/>Resource<!-- |
| − | * associate images to characters or character states (tables: Resource, ResourceVariant, BaseEntity.specific_rights_text should contain the corresponding picture’s author name like: “© Author Name”), size of images with maximal height 40px | + | --><br/>├ ResourceVariant <!-- |
| − | Finally make | + | --><br/>│ (e.g. with ResourceVariant_Enum.code='best_quality')<!-- |
| − | * tag "Artenquiz" means it is a quiz | + | --><br/>└ ResourceVariant<!-- |
| − | * tag "Public" means it will be | + | --><br/> (e.g. with ResourceVariant_Enum.code='tiny_sample')</code> |
| − | * tag "Identification" make the project | + | #* eventually associate geographic area (table: GeographicAreaProxy, GeographicArea_Taxon) |
| − | + | # Create character and character states (table Descriptor, CategoricalState), character tree (tables: DescriptorTree, DescriptorTreeNode) | |
| − | + | #* associate images to characters or character states (tables: Resource, ResourceVariant, BaseEntity.specific_rights_text should contain the corresponding picture’s author name like: “© Author Name”), size of images with maximal height 40px (height should precced) | |
| − | + | # Finally make the project publicly available: set the Project‘s tag (table: Tag) | |
| − | + | #* tag "Artenquiz" means it is a quiz | |
| − | + | #* tag "Public" means it will be publicly accessible | |
| − | + | #* tag "Identification" make the project available for identification (URL: …/mobile_bestimmung/, TODO: not yet implemented) | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 12:43, 12 February 2021
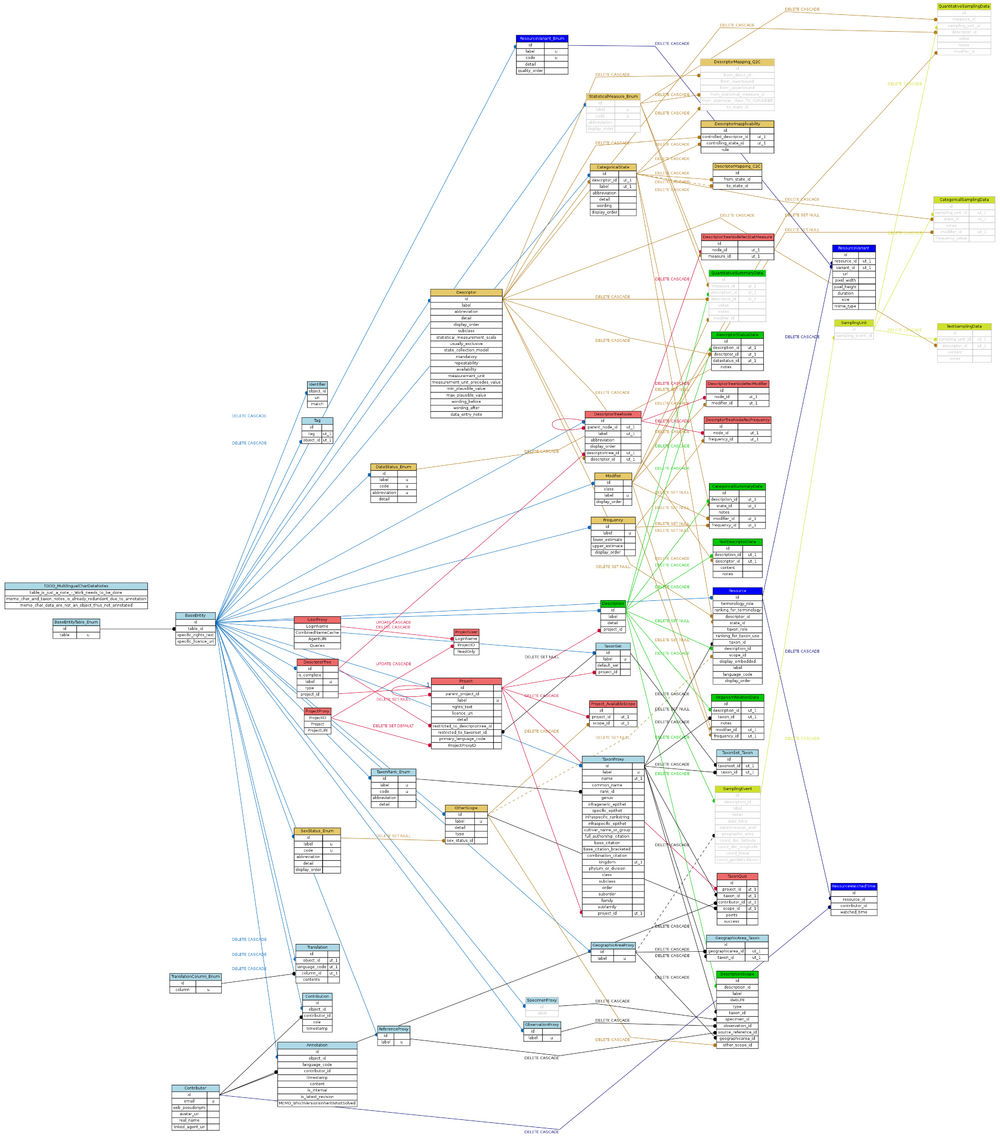
The ER diagram of DiversityDescriptions_Quiz (DD_Quiz) is a clone of DiversityDescriptions 3.00.10. July 17th, 2014 that is intended to work with the Web-Framework Django to have a quiz for species descriptions in German. This project is presently under development. Additional tables see below. For further information see under DiversityDescriptions Quiz implementations.
Suggested citation is that of DiversityDescriptions 3.00.10: G. Hagedorn, A. Plank, A. Link, G. Rambold & D. Triebel (2014). DiversityDescriptions information model (version 3.00.10). http://www.diversityworkbench.net/Portal/DescriptionsModel_v3.00.10.
Hover the mouse over a table to jump to the documentation (linked to DiversityDescriptions 3.00.10. July 17th, 2014).
Contents
Documentation
Database
Scheme of DiversityDescriptions Quiz:
Graph image creation requires permission to upload.
The following conventions and abbreviations have been used in the tables: Columns of primary key:underlined. Categorical data are presented to a user in the quiz therefore some tables (in gray) are not used for the quiz.
The data base scheme described in DOT language (click the expand on the right side to read the content)
<graphviz caption='Scheme of Diversity Descriptions Quiz' alt='Diversity Descriptions Quiz' format='png'>
digraph DD_Quiz_Scheme_20140717{
graph [rankdir = "LR" splines="line" outputorder="edgesfirst"];
node [fontsize = "10", shape = "plaintext"];
edge [color = "black" arrowhead="dot"];
Annotation [
URL="[[DescriptionsModel_v3.00.10#Table:_Annotation]]"
tooltip="One record per annotated object (Char, State, Descr. etc.)"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="1">Annotation</td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="object_id">object_id</td></tr>
<tr><td PORT="language_code">language_code</td></tr>
<tr><td PORT="contributor_id">contributor_id</td></tr>
<tr><td PORT="timestamp">timestamp</td></tr>
<tr><td PORT="content">content</td></tr>
<tr><td PORT="is_internal">is_internal</td></tr>
<tr><td PORT="is_latest_revision">is_latest_revision</td></tr>
<tr><td PORT="MEMO_WhichVersionsInheritIsNotSolved">MEMO_WhichVersionsInheritIsNotSolved</td></tr>
//CONSTRAINT [PK_Annotation] PRIMARY KEY ([id])
</table>>
];
BaseEntity [
URL="[[DescriptionsModel_v3.00.10#Table:_BaseEntity]]"
tooltip="The BaseEntity is used within the database to provide unique keys"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0">
<tr><td BGCOLOR="lightblue" >BaseEntity</td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="table_id">table_id</td></tr>
<tr><td PORT="specific_rights_text">specific_rights_text</td></tr>
<tr><td PORT="specific_licence_uri">specific_licence_uri</td></tr>
//CONSTRAINT [PK_BaseEntity] PRIMARY KEY ([id])
</table>>
];
BaseEntityTable_Enum [
URL="[[DescriptionsModel_v3.00.10#Table:_BaseEntityTable_Enum]]"
tooltip="The BaseEntityTable_Enum contains the names of tables that reference the BaseEntity table"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">BaseEntityTable_Enum</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="table">table</td><td>u</td></tr>
//CONSTRAINT [PK_BaseEntityTable_Enum] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_BaseEntityTable_Enum_1] UNIQUE ([table])
</table>>
];
CategoricalSamplingData [
URL="[[DescriptionsModel_v3.00.10#Table:_CategoricalSamplingData]]"
tooltip="The categorical data recorded for a sampling event"
fontcolor="gray"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray">
<tr><td BGCOLOR="#cee12c" colspan="2"><font color="black">CategoricalSamplingData</font></td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="sampling_unit_id">sampling_unit_id</td><td>ut_1</td></tr>
<tr><td PORT="state_id">state_id</td><td>ut_1</td></tr>
<tr><td PORT="notes">notes</td><td></td></tr>
<tr><td PORT="modifier_id">modifier_id</td><td>ut_1</td></tr>
<tr><td PORT="frequency_value">frequency_value</td><td></td></tr>
//CONSTRAINT [PK_CategoricalSamplingData] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_CategoricalSamplingData_1] UNIQUE ([sampling_unit_id],[state_id],[modifier_id])
</table>>
];
CategoricalState [
URL="[[DescriptionsModel_v3.00.10#Table:_CategoricalState]]"
tooltip="The categorical states available for categorical descriptors"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#e5c96a" colspan="2">CategoricalState</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="descriptor_id">descriptor_id</td><td>ut_1</td></tr>
<tr><td PORT="label">label</td><td>ut_1</td></tr>
<tr><td PORT="abbreviation">abbreviation</td><td></td></tr>
<tr><td PORT="detail">detail</td><td></td></tr>
<tr><td PORT="wording">wording</td><td></td></tr>
<tr><td PORT="display_order">display_order</td><td></td></tr>
//CONSTRAINT [PK_CategoricalState] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_CategoricalState_1] UNIQUE ([label],[descriptor_id])
</table>>
];
CategoricalSummaryData [
URL="[[DescriptionsModel_v3.00.10#Table:_CategoricalSummaryData]]"
tooltip="The categorical data of a description"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#00c900" colspan="2">CategoricalSummaryData</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="description_id">description_id</td><td>ut_1</td></tr>
<tr><td PORT="state_id">state_id</td><td>ut_1</td></tr>
<tr><td PORT="notes">notes</td><td></td></tr>
<tr><td PORT="modifier_id">modifier_id</td><td>ut_1</td></tr>
<tr><td PORT="frequency_id">frequency_id</td><td>ut_1</td></tr>
//CONSTRAINT [PK_CategoricalSummaryData] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_CategoricalSummaryData_1] UNIQUE ([description_id],[state_id],[modifier_id],[frequency_id])
</table>>
];
Contribution [
URL="[[#Table:_Contribution]]"
tooltip="Multiple contributors may contribute in different roles"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="1">Contribution</td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="object_id">object_id</td></tr>
<tr><td PORT="contributor_id">contributor_id</td></tr>
<tr><td PORT="role">role</td></tr>
<tr><td PORT="timestamp">timestamp</td></tr>
//CONSTRAINT [PK_Contribution] PRIMARY KEY ([id])
</table>>
];
Contributor [
URL="[[#Table:_Contributor]]"
tooltip="People contibuting to a work. Roles are recorded in Contribution"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">Contributor</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="email">email</td><td>u</td></tr>
<tr><td PORT="web_pseudonym">web_pseudonym</td><td></td></tr>
<tr><td PORT="avatar_uri">avatar_uri</td><td></td></tr>
<tr><td PORT="real_name">real_name</td><td></td></tr>
<tr><td PORT="linked_agent_uri">linked_agent_uri</td><td></td></tr>
//CONSTRAINT [PK_Contributor] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_Contributor_1] UNIQUE ([email])
</table>>
];
DataStatus_Enum [
URL="[[DescriptionsModel_v3.00.10#Table:_DataStatus_Enum]]"
tooltip="Values of data status used for descriptions according to SDD 1.1 rev 5"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#e5c96a" colspan="2">DataStatus_Enum</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="label">label</td><td>u</td></tr>
<tr><td PORT="code">code</td><td>u</td></tr>
<tr><td PORT="abbreviation">abbreviation</td><td>u</td></tr>
<tr><td PORT="detail">detail</td><td></td></tr>
//CONSTRAINT [PK_DataStatus_Enum] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_DataStatus_Enum_1] UNIQUE ([code]),
//CONSTRAINT [UQ_DataStatus_Enum_2] UNIQUE ([abbreviation]),
//CONSTRAINT [UQ_DataStatus_Enum_3] UNIQUE ([label])
</table>>
];
Description [
URL="[[DescriptionsModel_v3.00.10#Table:_Description]]"
tooltip="The description in the database"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#00c900" colspan="1">Description</td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="label">label</td></tr>
<tr><td PORT="detail">detail</td></tr>
<tr><td PORT="project_id">project_id</td></tr>
//CONSTRAINT [PK_Description] PRIMARY KEY ([id])
</table>>
];
DescriptionScope [
URL="[[DescriptionsModel_v3.00.10#Table:_DescriptionScope]]"
tooltip="The scope of the description"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#00c900" colspan="1">DescriptionScope</td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="description_id">description_id</td></tr>
<tr><td PORT="label">label</td></tr>
<tr><td PORT="dwbURI">dwbURI</td></tr>
<tr><td PORT="type">type</td></tr>
<tr><td PORT="taxon_id">taxon_id</td></tr>
<tr><td PORT="specimen_id">specimen_id</td></tr>
<tr><td PORT="observation_id">observation_id</td></tr>
<tr><td PORT="source_reference_id">source_reference_id</td></tr>
<tr><td PORT="geographicarea_id">geographicarea_id</td></tr>
<tr><td PORT="other_scope_id">other_scope_id</td></tr>
//CONSTRAINT [PK_DescriptionScope] PRIMARY KEY ([id])
</table>>
];
Descriptor [
URL="[[DescriptionsModel_v3.00.10#Table:_Descriptor]]"
tooltip="Descriptor (= characters, features) define variables"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#e5c96a">Descriptor</td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="label">label</td></tr>
<tr><td PORT="abbreviation">abbreviation</td></tr>
<tr><td PORT="detail">detail</td></tr>
<tr><td PORT="display_order">display_order</td></tr>
<tr><td PORT="subclass">subclass</td></tr>
<tr><td PORT="statistical_measurement_scale">statistical_measurement_scale</td></tr>
<tr><td PORT="usually_exclusive">usually_exclusive</td></tr>
<tr><td PORT="state_collection_model">state_collection_model</td></tr>
<tr><td PORT="mandatory">mandatory</td></tr>
<tr><td PORT="repeatability">repeatability</td></tr>
<tr><td PORT="availability">availability</td></tr>
<tr><td PORT="measurement_unit">measurement_unit</td></tr>
<tr><td PORT="measurement_unit_precedes_value">measurement_unit_precedes_value</td></tr>
<tr><td PORT="min_plausible_value">min_plausible_value</td></tr>
<tr><td PORT="max_plausible_value">max_plausible_value</td></tr>
<tr><td PORT="wording_before">wording_before</td></tr>
<tr><td PORT="wording_after">wording_after</td></tr>
<tr><td PORT="data_entry_note">data_entry_note</td></tr>
//CONSTRAINT [PK_Descriptor] PRIMARY KEY ([id])
</table>>
];
DescriptorInapplicability [
URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorInapplicability]]"
tooltip="The descriptor dependency rules"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#e5c96a" colspan="2">DescriptorInapplicability</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="controlled_descriptor_id">controlled_descriptor_id</td><td>ut_1</td></tr>
<tr><td PORT="controlling_state_id">controlling_state_id</td><td>ut_1</td></tr>
<tr><td PORT="rule">rule</td><td></td></tr>
//CONSTRAINT [PK_DescriptorInapplicability] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_DescriptorInapplicability_1] UNIQUE ([controlled_descriptor_id],[controlling_state_id])
</table>>
];
DescriptorMapping_C2C [
URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorMapping_C2C]]"
tooltip="Mapping of a categorical state to another categorical state"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#e5c96a" colspan="1">DescriptorMapping_C2C</td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="from_state_id">from_state_id</td></tr>
<tr><td PORT="to_state_id">to_state_id</td></tr>
//CONSTRAINT [PK_DescriptorMapping_C2C] PRIMARY KEY ([id])
</table>>
];
DescriptorMapping_Q2C [
URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorMapping_Q2C]]" fontcolor="gray"
tooltip="Mapping of a quantitative descriptor to a categorical state"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray">
<tr><td BGCOLOR="#e5c96a" colspan="1"><font color="black">DescriptorMapping_Q2C</font></td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="from_descr_id">from_descr_id</td></tr>
<tr><td PORT="from_lowerbound">from_lowerbound</td></tr>
<tr><td PORT="from_upperbound">from_upperbound</td></tr>
<tr><td PORT="from_statistical_measure_id">from_statistical_measure_id</td></tr>
<tr><td PORT="from_statmeas_class_TO_CONSIDER">from_statmeas_class_TO_CONSIDER</td></tr>
<tr><td PORT="to_state_id">to_state_id</td></tr>
//CONSTRAINT [PK_DescriptorMapping_Q2C] PRIMARY KEY ([id])
</table>>
];
DescriptorStatusData [
URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorStatusData]]"
tooltip="The status data of a descriptor for a certain description"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#00c900" colspan="2">DescriptorStatusData</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="description_id">description_id</td><td>ut_1</td></tr>
<tr><td PORT="descriptor_id">descriptor_id</td><td>ut_1</td></tr>
<tr><td PORT="datastatus_id">datastatus_id</td><td>ut_1</td></tr>
<tr><td PORT="notes">notes</td><td></td></tr>
//CONSTRAINT [PK_DescriptorStatusData] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_DescriptorStatusData_1] UNIQUE ([description_id],[descriptor_id],[datastatus_id])
</table>>
];
DescriptorTree [
URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorTree]]"
tooltip="The root and definition of a descriptor tree"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="2">DescriptorTree</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="is_complete">is_complete</td><td></td></tr>
<tr><td PORT="label">label</td><td>u</td></tr>
<tr><td PORT="type">type</td><td></td></tr>
<tr><td PORT="project_id">project_id</td><td PORT="project_id_e"></td></tr>
//CONSTRAINT [PK_DescriptorTree] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_DescriptorTree_1] UNIQUE ([label])
</table>>
];
DescriptorTreeNode [
URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorTreeNode]]"
tooltip="The descriptor tree nodes representing either nodes of the tree or descriptors ("leafes" of the tree)"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="2">DescriptorTreeNode</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="parent_node_id">parent_node_id</td><td PORT="parent_node_id_e">ut_1</td></tr>
<tr><td PORT="label">label</td><td PORT="label_e">ut_1</td></tr>
<tr><td PORT="abbreviation">abbreviation</td><td PORT="abbreviation_e"></td></tr>
<tr><td PORT="display_order">display_order</td><td PORT="display_order_e"></td></tr>
<tr><td PORT="descriptortree_id">descriptortree_id</td><td PORT="descriptortree_id_e">ut_1</td></tr>
<tr><td PORT="descriptor_id">descriptor_id</td><td PORT="descriptor_id_e">ut_1</td></tr>
//CONSTRAINT [PK_DescriptorTreeNode] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_DescriptorTreeNode_1] UNIQUE ([descriptortree_id],[parent_node_id],[descriptor_id],[label])
</table>>
];
DescriptorTreeNodeRecFrequency [
URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorTreeNodeRecFrequency]]"
tooltip="Selection of recommended frequency values for descriptor tree parts or single descriptors"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="2">DescriptorTreeNodeRecFrequency</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="node_id">node_id</td><td>ut_1</td></tr>
<tr><td PORT="frequency_id">frequency_id</td><td>ut_1</td></tr>
//CONSTRAINT [PK_DescriptorTreeNodeRecFrequency] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_DescriptorTreeNodeRecFrequency_1] UNIQUE ([node_id],[frequency_id])
</table>>
];
DescriptorTreeNodeRecModifier [
URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorTreeNodeRecModifier]]"
tooltip="Selection of recommended modifier values for descriptor tree parts or single descriptors"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="2">DescriptorTreeNodeRecModifier</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="node_id">node_id</td><td>ut_1</td></tr>
<tr><td PORT="modifier_id">modifier_id</td><td>ut_1</td></tr>
//CONSTRAINT [PK_DescriptorTreeNodeRecModifier] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_DescriptorTreeNodeRecModifier_1] UNIQUE ([node_id],[modifier_id])
</table>>
];
DescriptorTreeNodeRecStatMeasure [
URL="[[DescriptionsModel_v3.00.10#Table:_DescriptorTreeNodeRecStatMeasure]]"
tooltip="Selection of recommended statistical measures for descriptor tree parts or single descriptors"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="2">DescriptorTreeNodeRecStatMeasure</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="node_id">node_id</td><td>ut_1</td></tr>
<tr><td PORT="measure_id">measure_id</td><td>ut_1</td></tr>
//CONSTRAINT [PK_DescriptorTreeNodeRecStatMeasure] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_DescriptorTreeNodeRecStatMeasure_1] UNIQUE ([node_id],[measure_id])
</table>>
];
Frequency [
URL="[[DescriptionsModel_v3.00.10#Table:_Frequency]]"
tooltip="Definition of frequency modifier values"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#e5c96a" colspan="2">Frequency</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="label">label</td><td PORT="label_e">u</td></tr>
<tr><td PORT="lower_estimate">lower_estimate</td><td PORT="lower_estimate_e"></td></tr>
<tr><td PORT="upper_estimate">upper_estimate</td><td PORT="upper_estimate_e"></td></tr>
<tr><td PORT="display_order">display_order</td><td PORT="display_order_e"></td></tr>
//CONSTRAINT [PK_Frequency] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_Frequency_1] UNIQUE ([label])
</table>>
];
GeographicAreaProxy [
URL="[[DescriptionsModel_v3.00.10#Table:_GeographicAreaProxy]]"
tooltip="Geographic Area, proxy for WB component"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">GeographicAreaProxy</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="label">label</td><td PORT="label_e">u</td></tr>
//CONSTRAINT [PK_GeographicAreaProxy] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_GeographicAreaProxy_1] UNIQUE ([label])
</table>>
];
GeographicArea_Taxon [
URL="[[DescriptionsModel_v3.00.10#Table:_GeographicArea_Taxon]]"
tooltip="Presence (or likely presence) of taxon in a given geographic area"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">GeographicArea_Taxon</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="geographicarea_id">geographicarea_id</td><td>ut_1</td></tr>
<tr><td PORT="taxon_id">taxon_id</td><td>ut_1</td></tr>
//CONSTRAINT [PK_GeographicArea_Taxon] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_GeographicArea_Taxon_1] UNIQUE ([geographicarea_id],[taxon_id])
</table>>
];
Identifier [
URL="[[DescriptionsModel_v3.00.10#Table:_Identifier]]"
tooltip="Objects may have multiple external identifiers"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="1">Identifier</td></tr>
<tr><td PORT="object_id">object_id</td></tr>
<tr><td PORT="uri">uri</td></tr>
<tr><td PORT="match">match</td></tr>
//CONSTRAINT [PK_Identifier] PRIMARY KEY ([uri])
</table>>
];
Modifier [
URL="[[DescriptionsModel_v3.00.10#Table:_Modifier]]"
tooltip="Definition of modifier values"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#e5c96a" colspan="2">Modifier</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="class">class</td><td></td></tr>
<tr><td PORT="label">label</td><td>u</td></tr>
<tr><td PORT="display_order">display_order</td><td></td></tr>
//CONSTRAINT [PK_Modifier] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_Modifier_1] UNIQUE ([label])
</table>>
];
ObservationProxy [
URL="[[DescriptionsModel_v3.00.10#Table:_ObservationProxy]]"
tooltip="Observation; proxy for WB component"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">ObservationProxy</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="label">label</td><td PORT="label_e">u</td></tr>
//CONSTRAINT [PK_ObservationProxy] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_ObservationProxy_1] UNIQUE ([label])
</table>>
];
OrganismRelationData [
URL="[[DescriptionsModel_v3.00.10#Table:_OrganismRelationData]]"
tooltip="Categorical data (states recorded for a description)"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#00c900" colspan="2">OrganismRelationData</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="description_id">description_id</td><td>ut_1</td></tr>
<tr><td PORT="taxon_id">taxon_id</td><td>ut_1</td></tr>
<tr><td PORT="notes">notes</td><td></td></tr>
<tr><td PORT="modifier_id">modifier_id</td><td>ut_1</td></tr>
<tr><td PORT="frequency_id">frequency_id</td><td>ut_1</td></tr>
//CONSTRAINT [PK_OrganismRelationData] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_OrganismRelationData_1] UNIQUE ([description_id],[taxon_id],[modifier_id],[frequency_id])
</table>>
];
OtherScope [
URL="[[DescriptionsModel_v3.00.10#Table:_OtherScope]]"
tooltip="Scope values for description scope values for scope types 'Other scope', 'Part', 'Stage' and 'Sex' "
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#e5c96a" colspan="2">OtherScope</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="label">label</td><td PORT="label_e">u</td></tr>
<tr><td PORT="detail">detail</td><td PORT="detail_e"></td></tr>
<tr><td PORT="type">type</td><td PORT="type_e"></td></tr>
<tr><td PORT="sex_status_id">sex_status_id</td><td PORT="sex_status_id_e"></td></tr>
//CONSTRAINT [PK_OtherScope] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_OtherScope_1] UNIQUE ([label])
</table>>
];
Project [
URL="[[DescriptionsModel_v3.00.10#Table:_Project]]"
tooltip="Projects define separated workareas in a single database"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="2">Project</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="parent_project_id">parent_project_id</td><td PORT="parent_project_id_e"></td></tr>
<tr><td PORT="label">label</td><td PORT="label_e">u</td></tr>
<tr><td PORT="rights_text">rights_text</td><td PORT="rights_text_e"></td></tr>
<tr><td PORT="licence_uri">licence_uri</td><td PORT="licence_uri_e"></td></tr>
<tr><td PORT="detail">detail</td><td PORT="detail_e"></td></tr>
<tr><td PORT="restricted_to_descriptortree_id">restricted_to_descriptortree_id</td><td PORT="restricted_to_descriptortree_id_e"></td></tr>
<tr><td PORT="restricted_to_taxonset_id">restricted_to_taxonset_id</td><td PORT="restricted_to_taxonset_id_e"></td></tr>
<tr><td PORT="primary_language_code">primary_language_code</td><td PORT="primary_language_code_e"></td></tr>
<tr><td PORT="ProjectProxyID">ProjectProxyID</td><td PORT="ProjectProxyID_e"></td></tr>
//CONSTRAINT [PK_Project] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_Project_1] UNIQUE ([label])
</table>>
];
Project_AvailableScope [
URL="[[DescriptionsModel_v3.00.10#Table:_Project_AvailableScope]]"
tooltip="Scope values available for a certain project"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="2">Project_AvailableScope</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="project_id">project_id</td><td>ut_1</td></tr>
<tr><td PORT="scope_id">scope_id</td><td>ut_1</td></tr>
//CONSTRAINT [PK_Project_AvailableScope] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_Project_AvailableScope_1] UNIQUE ([project_id],[scope_id])
</table>>
];
QuantitativeSamplingData [
URL="[[DescriptionsModel_v3.00.10#Table:_QuantitativeSamplingData]]"
tooltip="The quantitative data recorded for a sampling event"
fontcolor="gray"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray">
<tr><td BGCOLOR="#cee12c" colspan="1"><font color="black">QuantitativeSamplingData</font></td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="measure_id">measure_id</td></tr>
<tr><td PORT="sampling_unit_id">sampling_unit_id</td></tr>
<tr><td PORT="descriptor_id">descriptor_id</td></tr>
<tr><td PORT="value">value</td></tr>
<tr><td PORT="notes">notes</td></tr>
<tr><td PORT="modifier_id">modifier_id</td></tr>
//CONSTRAINT [PK_QuantitativeSamplingData] PRIMARY KEY ([id])
</table>>
];
QuantitativeSummaryData [
URL="[[DescriptionsModel_v3.00.10#Table:_QuantitativeSummaryData]]"
tooltip="The quantitative data recorded for a sampling event" fontcolor="gray"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray">
<tr><td BGCOLOR="#00c900" colspan="2"><font color="black">QuantitativeSummaryData</font></td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="measure_id">measure_id</td><td>ut_1</td></tr>
<tr><td PORT="description_id">description_id</td><td>ut_1</td></tr>
<tr><td PORT="descriptor_id">descriptor_id</td><td>ut_1</td></tr>
<tr><td PORT="value">value</td><td></td></tr>
<tr><td PORT="notes">notes</td><td></td></tr>
<tr><td PORT="modifier_id">modifier_id</td><td></td></tr>
//CONSTRAINT [PK_QuantitativeSummaryData] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_QuantitativeSummaryData_1] UNIQUE ([description_id],[measure_id],[descriptor_id])
</table>>
];
ReferenceProxy [
URL="[[DescriptionsModel_v3.00.10#Table:_ReferenceProxy]]"
tooltip="Reference (literature/source); proxy for WB component"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">ReferenceProxy</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="label">label</td><td>u</td></tr>
//CONSTRAINT [PK_ReferenceProxy] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_ReferenceProxy_1] UNIQUE ([label])
</table>>
];
Resource [
URL="[[DescriptionsModel_v3.00.10#Table:_Resource]]"
tooltip="Hyperlinks to separate rich text/media objects"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="blue" colspan="1"><font COLOR="white">Resource</font></td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="terminology_role">terminology_role</td></tr>
<tr><td PORT="ranking_for_terminology">ranking_for_terminology</td></tr>
<tr><td PORT="descriptor_id">descriptor_id</td></tr>
<tr><td PORT="state_id">state_id</td></tr>
<tr><td PORT="taxon_role">taxon_role</td></tr>
<tr><td PORT="ranking_for_taxon_use">ranking_for_taxon_use</td></tr>
<tr><td PORT="taxon_id">taxon_id</td></tr>
<tr><td PORT="description_id">description_id</td></tr>
<tr><td PORT="scope_id">scope_id</td></tr>
<tr><td PORT="display_embedded">display_embedded</td></tr>
<tr><td PORT="label">label</td></tr>
<tr><td PORT="language_code">language_code</td></tr>
<tr><td PORT="display_order">display_order</td></tr>
//CONSTRAINT [PK_Resource] PRIMARY KEY ([id])
</table>>
];
ResourceVariant [
URL="[[DescriptionsModel_v3.00.10#Table:_ResourceVariant]]"
tooltip="Different resource variants/instances/service access points"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="blue" colspan="2"><font COLOR="white">ResourceVariant</font></td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="resource_id">resource_id</td><td>ut_1</td></tr>
<tr><td PORT="variant_id">variant_id</td><td>ut_1</td></tr>
<tr><td PORT="url">url</td><td></td></tr>
<tr><td PORT="pixel_width">pixel_width</td><td></td></tr>
<tr><td PORT="pixel_height">pixel_height</td><td></td></tr>
<tr><td PORT="duration">duration</td><td></td></tr>
<tr><td PORT="size">size</td><td></td></tr>
<tr><td PORT="mime_type">mime_type</td><td></td></tr>
//CONSTRAINT [PK_ResourceVariant] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_ResourceVariant_1] UNIQUE ([resource_id],[variant_id])
</table>>
];
ResourceVariant_Enum [
URL="[[DescriptionsModel_v3.00.10#Table:_ResourceVariantEnum]]"
tooltip="Classes for resource variants, values are predefined in the database"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="blue" colspan="2"><font COLOR="white">ResourceVariant_Enum</font></td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="label">label</td><td PORT="label_e">u</td></tr>
<tr><td PORT="code">code</td><td PORT="code_e">u</td></tr>
<tr><td PORT="detail">detail</td><td PORT="detail_e"></td></tr>
<tr><td PORT="quality_order">quality_order</td><td PORT="quality_order_e"></td></tr>
//CONSTRAINT [PK_ResourceVariant_Enum] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_ResourceVariant_Enum_1] UNIQUE ([label]),
//CONSTRAINT [UQ_ResourceVariant_Enum_2] UNIQUE ([code])
</table>>
];
ResourceWatchedTime [
URL="[[#Table:_ResourceWatchedTime]]"
tooltip="The time a resource thumb was watched (=mouse hover)"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="blue" colspan="1"><font COLOR="white">ResourceWatchedTime</font></td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="resource_id">resource_id</td></tr>
<tr><td PORT="contributor_id">contributor_id</td></tr>
<tr><td PORT="watched_time">watched_time</td></tr>
</table>>
];
SamplingEvent [
URL="[[DescriptionsModel_v3.00.10#Table:_SamplingEvent]]"
tooltip="A sampling event may contain many sampling units" fontcolor="gray"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray">
<tr><td BGCOLOR="#cee12c" colspan="1"><font color="black">SamplingEvent</font></td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="description_id">description_id</td></tr>
<tr><td PORT="label">label</td></tr>
<tr><td PORT="detail">detail</td></tr>
<tr><td PORT="date_time">date_time</td></tr>
<tr><td PORT="datetimespan_end">datetimespan_end</td></tr>
<tr><td PORT="geographic_area">geographic_area</td></tr>
<tr><td PORT="coord_dec_latitude">coord_dec_latitude</td></tr>
<tr><td PORT="coord_dec_longitude">coord_dec_longitude</td></tr>
<tr><td PORT="coord_literal">coord_literal</td></tr>
<tr><td PORT="coord_geodeticdatum">coord_geodeticdatum</td></tr>
//CONSTRAINT [PK_SamplingEvent] PRIMARY KEY ([id])
</table>>
];
SamplingUnit [
URL="[[DescriptionsModel_v3.00.10#Table:_SamplingUnit]]"
tooltip="Categorical data" fontcolor="gray"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COlOR="gray">
<tr><td BGCOLOR="#cee12c" colspan="1"><font color="black">SamplingUnit</font></td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="sampling_event_id">sampling_event_id</td></tr>
//CONSTRAINT [PK_SamplingUnit] PRIMARY KEY ([id])
</table>>
];
SexStatus_Enum [
URL="[[DescriptionsModel_v3.00.10#Table:_SexStatus_Enum]]"
tooltip="Values of sex status predefined according to SDD 1.1 rev 5"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#e5c96a" colspan="2">SexStatus_Enum</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="label">label</td><td PORT="label_e">u</td></tr>
<tr><td PORT="code">code</td><td PORT="code_e">u</td></tr>
<tr><td PORT="abbreviation">abbreviation</td><td PORT="abbreviation_e"></td></tr>
<tr><td PORT="detail">detail</td><td PORT="detail_e"></td></tr>
<tr><td PORT="display_order">display_order</td><td PORT="display_order_e"></td></tr>
//CONSTRAINT [PK_SexStatus_Enum] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_SexStatus_Enum_1] UNIQUE ([label]),
//CONSTRAINT [UQ_SexStatus_Enum_2] UNIQUE ([code])
</table>>
];
SpecimenProxy [
URL="[[DescriptionsModel_v3.00.10#Table:_SpecimenProxy]]"
tooltip="Specimens from collections; proxy for WB component" fontcolor="gray"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray">
<tr><td BGCOLOR="lightblue" colspan="1"><font color="black">SpecimenProxy</font></td></tr>
<tr><td PORT="id">id</td></tr>
<tr><td PORT="label">label</td>u</tr>
//CONSTRAINT [PK_SpecimenProxy] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_SpecimenProxy_1] UNIQUE ([label])
</table>>
];
StatisticalMeasure_Enum [
URL="[[DescriptionsModel_v3.00.10#Table:_StatisticalMeasure_Enum]]" fontcolor="gray"
tooltip="The statistical measures predefined according SDD 1.1 rev 5"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray">
<tr><td BGCOLOR="#e5c96a" colspan="2"><font color="black">StatisticalMeasure_Enum</font></td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="label">label</td><td>u</td></tr>
<tr><td PORT="code">code</td><td>u</td></tr>
<tr><td PORT="abbreviation">abbreviation</td><td></td></tr>
<tr><td PORT="display_order">display_order</td><td></td></tr>
//CONSTRAINT [PK_StatisticalMeasure_Enum] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_StatisticalMeasure_Enum_1] UNIQUE ([label]),
//CONSTRAINT [UQ_StatisticalMeasure_Enum_2] UNIQUE ([code])
</table>>
];
Tag [
URL="[[DescriptionsModel_v3.00.10#Table:_Tag]]"
tooltip="Objects may have multiple tags (= keywords)"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">Tag</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="tag">tag</td><td>ut_1</td></tr>
<tr><td PORT="object_id">object_id</td><td>ut_1</td></tr>
//CONSTRAINT [PK_Tag] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_Tag_1] UNIQUE ([tag],[object_id])
</table>>
];
TaxonProxy [
URL="[[DescriptionsModel_v3.00.10#Table:_TaxonProxy]]"
tooltip="Taxon/tax. hierarchy (simplified); proxy for WB component"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">TaxonProxy</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="label">label</td><td>u</td></tr>
<tr><td PORT="name">name</td><td>ut_1</td></tr>
<tr><td PORT="common_name">common_name</td><td></td></tr>
<tr><td PORT="rank_id">rank_id</td><td></td></tr>
<tr><td PORT="genus">genus</td><td></td></tr>
<tr><td PORT="infrageneric_epithet">infrageneric_epithet</td><td></td></tr>
<tr><td PORT="specific_epithet">specific_epithet</td><td></td></tr>
<tr><td PORT="infraspecific_rankstring">infraspecific_rankstring</td><td></td></tr>
<tr><td PORT="infraspecific_epithet">infraspecific_epithet</td><td></td></tr>
<tr><td PORT="cultivar_name_or_group">cultivar_name_or_group</td><td></td></tr>
<tr><td PORT="full_authorship_citation">full_authorship_citation</td><td></td></tr>
<tr><td PORT="base_citation">base_citation</td><td></td></tr>
<tr><td PORT="base_citation_bracketed">base_citation_bracketed</td><td></td></tr>
<tr><td PORT="combination_citation">combination_citation</td><td></td></tr>
<tr><td PORT="kingdom">kingdom</td><td>ut_1</td></tr>
<tr><td PORT="phylum_or_division">phylum_or_division</td><td></td></tr>
<tr><td PORT="class">class</td><td></td></tr>
<tr><td PORT="subclass">subclass</td><td></td></tr>
<tr><td PORT="order">order</td><td></td></tr>
<tr><td PORT="suborder">suborder</td><td></td></tr>
<tr><td PORT="family">family</td><td></td></tr>
<tr><td PORT="subfamily">subfamily</td><td></td></tr>
<tr><td PORT="project_id">project_id</td><td>ut_1</td></tr>
//CONSTRAINT [PK_TaxonProxy] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_TaxonProxy_1] UNIQUE ([label]),
//CONSTRAINT [UQ_TaxonProxy_2] UNIQUE ([name],[kingdom],[project_id])
</table>>
];
TaxonRank_Enum [
URL="[[DescriptionsModel_v3.00.10#Table:_TaxonRank_Enum]]"
tooltip="Ranks for taxa (species, genus, family, order, class, etc.)"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">TaxonRank_Enum</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="label">label</td><td PORT="label_e">u</td></tr>
<tr><td PORT="code">code</td><td PORT="code_e">u</td></tr>
<tr><td PORT="abbreviation">abbreviation</td><td PORT="abbreviation_e"></td></tr>
<tr><td PORT="detail">detail</td><td PORT="detail_e"></td></tr>
//CONSTRAINT [PK_TaxonRank_Enum] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_TaxonRank_Enum_1] UNIQUE ([label]),
//CONSTRAINT [UQ_TaxonRank_Enum_2] UNIQUE ([code])
</table>>
];
TaxonSet [
URL="[[DescriptionsModel_v3.00.10#Table:_TaxonSet]]"
tooltip="Groups (or sets) of taxa; including non-phylogenetic groups "
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">TaxonSet</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="label">label</td><td PORT="label_e">u</td></tr>
<tr><td PORT="default_set">default_set</td><td PORT="default_set_e"></td></tr>
<tr><td PORT="project_id">project_id</td><td PORT="project_id_e"></td></tr>
//CONSTRAINT [PK_TaxonSet] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_TaxonSet_1] UNIQUE ([label])
</table>>
];
TaxonSet_Taxon [
URL="[[DescriptionsModel_v3.00.10#Table:_TaxonSet_Taxon]]"
tooltip="Membership of taxa in a taxon set (n:m relation)"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">TaxonSet_Taxon</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="taxonset_id">taxonset_id</td><td>ut_1</td></tr>
<tr><td PORT="taxon_id">taxon_id</td><td>ut_1</td></tr>
//CONSTRAINT [PK_TaxonSet_Taxon] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_TaxonSet_Taxon_1] UNIQUE ([taxonset_id],[taxon_id])
</table>>
];
TextDescriptorData [
URL="[[DescriptionsModel_v3.00.10#Table:_TextDescriptorData]]"
tooltip="The text data of a description"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#00c900" colspan="2">TextDescriptorData</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="description_id">description_id</td><td>ut_1</td></tr>
<tr><td PORT="descriptor_id">descriptor_id</td><td>ut_1</td></tr>
<tr><td PORT="content">content</td><td></td></tr>
<tr><td PORT="notes">notes</td><td></td></tr>
//CONSTRAINT [PK_TextDescriptorData] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_TextDescriptorData_1] UNIQUE ([description_id],[descriptor_id])
</table>>
];
TextSamplingData [
URL="[[DescriptionsModel_v3.00.10#Table:_TextSamplingData]]"
tooltip="Free-form text data recorded for a sampling event" fontcolor="gray"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" COLOR="gray">
<tr><td BGCOLOR="#cee12c" colspan="2"><font color="black">TextSamplingData</font></td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="sampling_unit_id">sampling_unit_id</td><td>ut_1</td></tr>
<tr><td PORT="descriptor_id">descriptor_id</td><td>ut_1</td></tr>
<tr><td PORT="content">content</td><td></td></tr>
<tr><td PORT="notes">notes</td><td></td></tr>
//CONSTRAINT [PK_TextSamplingData] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_TextSamplingData_1] UNIQUE ([sampling_unit_id],[descriptor_id])
</table>>
];
TODO_MultilingualCharDataNotes [
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="1">TODO_MultilingualCharDataNotes</td></tr>
<tr><td PORT="table_is_just_a_note_-_Work_needs_to_be_done">table_is_just_a_note_-_Work_needs_to_be_done</td></tr>
<tr><td PORT="memo_char_and_taxon_notes_is_already_redundant_due_to_annotation">memo_char_and_taxon_notes_is_already_redundant_due_to_annotation</td></tr>
<tr><td PORT="memo_char_data_are_not_an_object_thus_not_annotated">memo_char_data_are_not_an_object_thus_not_annotated</td></tr>
//CONSTRAINT [PK_TODO_MultilingualCharDataNotes] PRIMARY KEY ([table_is_just_a_note_-_Work_needs_to_be_done])
</table>>
];
Translation [
URL="[[DescriptionsModel_v3.00.10#Table:_Translation]]"
tooltip="The translations of entries related to BaseEntity"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">Translation</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="object_id">object_id</td><td>ut_1</td></tr>
<tr><td PORT="language_code">language_code</td><td>ut_1</td></tr>
<tr><td PORT="column_id">column_id</td><td>ut_1</td></tr>
<tr><td PORT="contents">contents</td><td></td></tr>
//CONSTRAINT [PK_Translation] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_Translation_1] UNIQUE ([object_id],[language_code],[column_id])
</table>>
];
TranslationColumn_Enum [
URL="[[DescriptionsModel_v3.00.10#Table:_TranslationColumn_Enum]]"
tooltip="The TranslationColumn_Enum contains the column names that are translated in the Translation table"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="lightblue" colspan="2">TranslationColumn_Enum</td></tr>
<tr><td PORT="id">id</td><td PORT="id_e"></td></tr>
<tr><td PORT="column">column</td><td PORT="column_e">u</td></tr>
//CONSTRAINT [PK_TranslationColumn_Enum] PRIMARY KEY ([id]),
//CONSTRAINT [UQ_TranslationColumn_Enum_1] UNIQUE ([column])
</table>>
];
ProjectProxy [
URL="[[DescriptionsModel_v3.00.10#Table:_ProjectProxy]]"
tooltip="The projects as stored in the module DiversityProjects"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="1">ProjectProxy</td></tr>
<tr><td PORT="ProjectID">ProjectID</td></tr>
<tr><td PORT="Project">Project</td></tr>
<tr><td PORT="ProjectURI">ProjectURI</td></tr>
//CONSTRAINT [PK_DiversityProjectProxy] PRIMARY KEY CLUSTERED ([ProjectID] ASC) WITH (PAD_INDEX = OFF, STATISTICS_NORECOMPUTE = OFF, IGNORE_DUP_KEY = OFF, ALLOW_ROW_LOCKS = ON, ALLOW_PAGE_LOCKS = ON, FILLFACTOR = 90) ON [PRIMARY]
</table>>
];
UserProxy [
URL="[[DescriptionsModel_v3.00.10#Table:_UserProxy]]"
tooltip="The user as stored in the module DiversityAgents"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="1">UserProxy</td></tr>
<tr><td PORT="LoginName">LoginName</td></tr>
<tr><td PORT="CombinedNameCache">CombinedNameCache</td></tr>
<tr><td PORT="AgentURI">AgentURI</td></tr>
<tr><td PORT="Queries">Queries</td></tr>
//CONSTRAINT [PK_UserProxy] PRIMARY KEY CLUSTERED ([LoginName] ASC) WITH (PAD_INDEX = OFF, STATISTICS_NORECOMPUTE = OFF, IGNORE_DUP_KEY = OFF, ALLOW_ROW_LOCKS = ON, ALLOW_PAGE_LOCKS = ON, FILLFACTOR = 90) ON [PRIMARY]
</table>>
];
ProjectUser [
URL="[[DescriptionsModel_v3.00.10#Table:_ProjectUser]]"
tooltip="The projects that a user can access"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="1">ProjectUser</td></tr>
<tr><td PORT="LoginName">LoginName</td></tr>
<tr><td PORT="ProjectID">ProjectID</td></tr>
<tr><td PORT="ReadOnly">ReadOnly</td></tr>
//CONSTRAINT [PK_ProjectUser] PRIMARY KEY CLUSTERED ([LoginName] ASC, [ProjectID])WITH (PAD_INDEX = OFF, STATISTICS_NORECOMPUTE = OFF, IGNORE_DUP_KEY = OFF, ALLOW_ROW_LOCKS = ON, ALLOW_PAGE_LOCKS = ON, FILLFACTOR = 90) ON [PRIMARY]
</table>>
];
TaxonQuiz [
URL="[[#Table:_TaxonQuiz]]"
tooltip="User’s correctly answered taxa"
label=<<table BORDER="0" BGCOLOR="white" CELLBORDER="1" CELLSPACING="0" >
<tr><td BGCOLOR="#ef6a6a" colspan="2">TaxonQuiz</td></tr>
<tr><td PORT="id">id</td><td></td></tr>
<tr><td PORT="project_id">project_id</td><td>ut_1</td></tr>
<tr><td PORT="taxon_id">taxon_id</td><td>ut_1</td></tr>
<tr><td PORT="contributor_id">contributor_id</td><td>ut_1</td></tr>
<tr><td PORT="scope_id">scope_id</td><td>ut_1</td></tr>
<tr><td PORT="points">points</td><td></td></tr>
<tr><td PORT="success">success</td><td></td></tr>
</table>>
];
BaseEntity:id:e -> Annotation:object_id:w[label="DELETE CASCADE" fontsize=10 color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> CategoricalState:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> Contribution:object_id:w[label="DELETE CASCADE" fontsize=10 color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> DataStatus_Enum:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> Description:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> Descriptor:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> DescriptorTree:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> DescriptorTreeNode:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> Frequency:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> GeographicAreaProxy:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> Identifier:object_id:w[label="DELETE CASCADE" fontsize=10 color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> Modifier:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> ObservationProxy:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> OtherScope:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> Project:id:w[color="#0069ba" fontcolor="#0069ba" headlabel="1" arrowhead="none"];
BaseEntity:id:e -> ReferenceProxy:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> Resource:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> ResourceVariant_Enum:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> SexStatus_Enum:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> SpecimenProxy:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> StatisticalMeasure_Enum:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> Tag:object_id:w[label="DELETE CASCADE" fontsize=10 color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> TaxonProxy:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> TaxonRank_Enum:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> TaxonSet:id:w[color="#0069ba" fontcolor="#0069ba"];
BaseEntity:id:e -> Translation:object_id:w[label="DELETE CASCADE" fontsize=10 color="#0069ba" fontcolor="#0069ba"];
BaseEntityTable_Enum:id -> BaseEntity:table_id;
CategoricalState:id_e:e -> CategoricalSamplingData:state_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
CategoricalState:id_e:e -> CategoricalSummaryData:state_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
CategoricalState:id_e:e -> DescriptorInapplicability:controlling_state_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
CategoricalState:id_e:e -> DescriptorMapping_C2C:from_state_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
CategoricalState:id_e:e -> DescriptorMapping_C2C:to_state_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817" style="dashed"];
CategoricalState:id_e:e -> DescriptorMapping_Q2C:to_state_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
CategoricalState:id_e:e -> Resource:state_id:w[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Contributor:id_e:e -> Annotation:contributor_id;
Contributor:id_e:e -> Contribution:contributor_id;
DataStatus_Enum:id -> DescriptorStatusData:datastatus_id[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Description:id:e -> CategoricalSummaryData:description_id[label="DELETE CASCADE" fontsize=10 color="#00c900" fontcolor="#00c900"];
Description:id:e -> DescriptionScope:description_id:w[label="DELETE CASCADE" fontsize=10 color="#00c900" fontcolor="#00c900"];
Description:id:e -> DescriptorStatusData:description_id[label="DELETE CASCADE" fontsize=10 color="#00c900" fontcolor="#00c900"];
Description:id:e -> OrganismRelationData:description_id[label="DELETE CASCADE" fontsize=10 color="#00c900" fontcolor="#00c900"];
Description:id:e -> QuantitativeSummaryData:description_id[label="DELETE CASCADE" fontsize=10 color="#00c900" fontcolor="#00c900"];
Description:id:e -> Resource:description_id:w[label="DELETE SET NULL" fontsize=10 color="#00c900" fontcolor="#00c900"];
Description:id:e -> SamplingEvent:description_id:w[label="DELETE CASCADE" fontsize=10 color="#00c900" fontcolor="#00c900"];
Description:id:e -> TextDescriptorData:description_id[label="DELETE CASCADE" fontsize=10 color="#00c900" fontcolor="#00c900"];
Descriptor:id:e -> CategoricalState:descriptor_id:w[color="#AF7817" fontcolor="#AF7817"];
Descriptor:id:e -> DescriptorInapplicability:controlled_descriptor_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Descriptor:id:e -> DescriptorMapping_Q2C:from_descr_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Descriptor:id:e -> DescriptorStatusData:descriptor_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Descriptor:id:e -> DescriptorTreeNode:descriptor_id:w[color="#AF7817" fontcolor="#AF7817"];
Descriptor:id:e -> QuantitativeSamplingData:descriptor_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Descriptor:id:e -> QuantitativeSummaryData:descriptor_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Descriptor:id:e -> Resource:descriptor_id:w[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Descriptor:id:e -> TextDescriptorData:descriptor_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Descriptor:id:e -> TextSamplingData:descriptor_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
DescriptorTree:id_e:e -> DescriptorTreeNode:descriptortree_id:w[color="#CC0033" fontcolor="#CC0033"];
DescriptorTree:id_e:e -> Project:restricted_to_descriptortree_id[label="DELETE SET NULL" fontsize=10 color="#CC0033" fontcolor="#CC0033"];
DescriptorTreeNode:id_e:e -> DescriptorTreeNodeRecFrequency:node_id[label="DELETE CASCADE" fontsize=10 color="#CC0033" fontcolor="#CC0033"];
DescriptorTreeNode:id_e:e -> DescriptorTreeNodeRecModifier:node_id[label="DELETE CASCADE" fontsize=10 color="#CC0033" fontcolor="#CC0033"];
DescriptorTreeNode:id_e:e -> DescriptorTreeNodeRecStatMeasure:node_id[label="DELETE CASCADE" fontsize=10 color="#CC0033" fontcolor="#CC0033"];
DescriptorTreeNode:id:w -> DescriptorTreeNode:parent_node_id:w[color="#CC0033" fontcolor="#CC0033"];
Frequency:id_e:e -> CategoricalSummaryData:frequency_id[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Frequency:id_e:e -> DescriptorTreeNodeRecFrequency:frequency_id[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Frequency:id_e:e -> OrganismRelationData:frequency_id[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
GeographicAreaProxy:id_e:e -> DescriptionScope:geographicarea_id:w[label="DELETE CASCADE" fontsize=10];
GeographicAreaProxy:id_e:e -> GeographicArea_Taxon:geographicarea_id[label="DELETE CASCADE" fontsize=10];
GeographicAreaProxy:id_e:e -> SamplingEvent:geographic_area[label="DELETE CASCADE" fontsize=10 style="dashed"];
Modifier:id_e:e -> CategoricalSamplingData:modifier_id:w[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Modifier:id_e:e -> CategoricalSummaryData:modifier_id[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Modifier:id_e:e -> DescriptorTreeNodeRecModifier:modifier_id[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Modifier:id_e:e -> OrganismRelationData:modifier_id[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Modifier:id_e:e -> QuantitativeSamplingData:modifier_id:w[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
Modifier:id_e:e -> QuantitativeSummaryData:modifier_id[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
ObservationProxy:id_e:e -> DescriptionScope:observation_id:w[label="DELETE CASCADE" fontsize=10];
OtherScope:id_e:e -> DescriptionScope:other_scope_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
OtherScope:id_e:e -> Project_AvailableScope:scope_id[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
OtherScope:id_e:e -> Resource:scope_id:w[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817" style="dashed"];
Project:id_e:e -> Description:project_id[color="#CC0033" fontcolor="#CC0033"];
Project:id:w -> DescriptorTree:project_id_e:w[color="#CC0033" fontcolor="#CC0033"];
Project:id_e:e -> Project_AvailableScope:project_id[label="DELETE CASCADE" fontsize=10 color="#CC0033" fontcolor="#CC0033"];
Project:id_e:e -> TaxonProxy:project_id[color="#CC0033" fontcolor="#CC0033"];
Project:id_e:e -> TaxonSet:project_id[color="#CC0033" fontcolor="#CC0033"];
Project:id:w -> Project:parent_project_id:w[color="#CC0033" fontcolor="#CC0033"];
ProjectProxy:ProjectID -> Project:ProjectProxyID[label="DELETE SET DEFAULT" fontsize=10 color="#CC0033" fontcolor="#CC0033"];
ProjectProxy:ProjectID -> ProjectUser:ProjectID[label="UPDATE CASCADE" fontsize=10 color="#CC0033" fontcolor="#CC0033"];
ReferenceProxy:id -> DescriptionScope:source_reference_id:w[label="DELETE CASCADE" fontsize=10];
Resource:id -> ResourceVariant:resource_id:w[label="DELETE CASCADE" fontsize=10 color="#000074" fontcolor="#000074"];
ResourceVariant_Enum:id_e:e -> ResourceVariant:variant_id:w[label="DELETE CASCADE" fontsize=10 color="#000074" fontcolor="#000074"];
SamplingEvent:id:e -> SamplingUnit:sampling_event_id:w[label="DELETE CASCADE" fontsize=10 fontcolor="#cee12c" color="#cee12c"];
SamplingUnit:id -> CategoricalSamplingData:sampling_unit_id:w[label="DELETE CASCADE" fontsize=10 fontcolor="#cee12c" color="#cee12c"];
SamplingUnit:id -> QuantitativeSamplingData:sampling_unit_id:w[label="DELETE CASCADE" fontsize=10 fontcolor="#cee12c" color="#cee12c"];
SamplingUnit:id -> TextSamplingData:sampling_unit_id[label="DELETE CASCADE" fontsize=10 fontcolor="#cee12c" color="#cee12c"];
SexStatus_Enum:id_e:e -> OtherScope:sex_status_id[label="DELETE SET NULL" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
SpecimenProxy:id -> DescriptionScope:specimen_id:w[label="DELETE CASCADE" fontsize=10];
StatisticalMeasure_Enum:id_e:e -> DescriptorMapping_Q2C:from_statistical_measure_id[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
StatisticalMeasure_Enum:id_e:e -> DescriptorTreeNodeRecStatMeasure:measure_id[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
StatisticalMeasure_Enum:id_e:e -> QuantitativeSamplingData:measure_id:w[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
StatisticalMeasure_Enum:id_e:e -> QuantitativeSummaryData:measure_id[label="DELETE CASCADE" fontsize=10 color="#AF7817" fontcolor="#AF7817"];
TaxonProxy:id_e:e -> DescriptionScope:taxon_id:w[label="DELETE CASCADE" fontsize=10];
TaxonProxy:id_e:e -> GeographicArea_Taxon:taxon_id[label="DELETE CASCADE" fontsize=10];
TaxonProxy:id_e:e -> OrganismRelationData:taxon_id[label="DELETE CASCADE" fontsize=10];
TaxonProxy:id_e:e -> Resource:taxon_id:w[label="DELETE SET NULL" fontsize=10];
TaxonProxy:id_e:e -> TaxonSet_Taxon:taxon_id[label="DELETE CASCADE" fontsize=10];
TaxonRank_Enum:id_e:e -> TaxonProxy:rank_id:w;
TaxonSet:id -> Project:restricted_to_taxonset_id_e[label="DELETE SET NULL" fontsize=10];
TaxonSet:id_e:e -> TaxonSet_Taxon:taxonset_id[label="DELETE CASCADE" fontsize=10];
TranslationColumn_Enum:id_e:e -> Translation:column_id;
UserProxy:LoginName -> ProjectUser:LoginName[label="UPDATE CASCADE\nDELETE CASCADE" fontsize=10 color="#CC0033" fontcolor="#CC0033"];
TaxonProxy:id_e:e -> TaxonQuiz:taxon_id:w
Contributor:id_e:e -> TaxonQuiz:contributor_id:w
OtherScope:id_e:e -> TaxonQuiz:scope_id:w
Project:id_e:e -> TaxonQuiz:project_id:w[color="#CC0033" fontcolor="#CC0033"];
Resource:id -> ResourceWatchedTime:resource_id:w[label="DELETE CASCADE" fontsize=10 color="#000074" fontcolor="#000074"]
Contributor:id_e:e -> ResourceWatchedTime:contributor_id:w[label="DELETE CASCADE" fontsize=10 color="#000074" fontcolor="#000074"]
}
</graphviz>
Table: Contribution
Multiple contributors may contribute in different roles
| Column | Data type | Description |
|---|---|---|
| id | int | Database-internal ID of this record (primary key) |
| object_id | int | Reference (foreign key) to a BaseEntity |
| contributor_id | int | Reference (foreign key) to a contributor |
| role | nvarchar (255) | one of 'creator', 'contributor', 'editor', 'photographer' |
| timestamp | datetime | Time stamp of the contribution |
Table: Contributor
People contibuting to a work. Roles are recorded in Contribution
| Column | Data type | Description |
|---|---|---|
| id | int | Database-internal ID of this record (primary key) |
| avatar_uri | nvarchar (255) | Optional URI to an image representing the user (default: NULL) |
| nvarchar (255) | User’s email address (default: NULL) | |
| linked_agent_uri | nvarchar (255) | Linking to agents managed in an external component (default: NULL) |
| real_name | nvarchar (255) | Optional name the user gave (full name) (default: NULL) |
| web_pseudonym | nvarchar (255) | Optional alternative name to display publicly on the web (default: NULL) |
Note: in the Quiz all users/contributors are requested to input their full name.
Table: ResourceWatchedTime
The time a resource thumb was watched (=mouse hover)
| Column | Data type | Description |
|---|---|---|
| id | int | Database-internal ID of this record (primary key) |
| resource_id | int | Reference (foreign key) to a resource |
| contributor_id | int | Reference (foreign key) to a contributor |
| watched_time | int | watched time in milliseconds |
Table: TaxonQuiz
User’s correctly answered taxa.
- unique together = ('project_id', 'contributor_id', 'taxon_id', 'scope_id')
| Column | Data type | Description |
|---|---|---|
| id | int | Database-internal ID of this record (primary key) |
| taxon_id | int | Reference (foreign kex) to a taxon that has been answerd correctly |
| project_id | int | Reference (foreign key) to the project |
| contributor_id | int | Reference (foreign key) to a contributor |
| scope_id | int | Reference to a scope (default: 0) |
| points | int | Points through scoring characters, already 3 added if success (default: 0) |
| sucess | tinyint | Was the taxon answered correctly by the user? (default: 0) |
New projects
Schematic important relations for a project:
Project ├ code (used for URL) ├ label (directly used for user display and in text context) ├ TaxonSet (i.e. taxa, media-resources etc.) ├ DescritorTree (i.e. characters and its states, media-resources etc.) ├ … …
Steps to enter data for a new project:
- Create a project (table: Project)
- Project.code is important: it is used as URL part and within the Django application as internal project identifier it should never be changed
- eventually create or associate scopes, like “female”, “male” (table: OtherScope, Project_AvailableScope)
- Eventually create the taxa (table: TaxonProxy)
- associate taxa to the project (tables: TaxonSet, TaxonSet_Taxon)
- associate images of taxa (tables: Resource, ResourceVariant, BaseEntity.specific_rights_text should contain the corresponding picture’s author name like: “© Author Name”). A set of maximum of 16 thumbs will be displayed
- an image is associated correctly, when 1 taxon has 1 Resource, but 2 (or multiple) ResourceVariant(!!)
- resource variants as ReourceVariant_Enum.code "tiny_sample" (=thumb 75×75px) and "best_quality" (max 500×500px displayed) should have corresponding urls in ResourceVariant.url:
Resource
├ ResourceVariant
│ (e.g. with ResourceVariant_Enum.code='best_quality')
└ ResourceVariant
(e.g. with ResourceVariant_Enum.code='tiny_sample')
- eventually associate geographic area (table: GeographicAreaProxy, GeographicArea_Taxon)
- Create character and character states (table Descriptor, CategoricalState), character tree (tables: DescriptorTree, DescriptorTreeNode)
- associate images to characters or character states (tables: Resource, ResourceVariant, BaseEntity.specific_rights_text should contain the corresponding picture’s author name like: “© Author Name”), size of images with maximal height 40px (height should precced)
- Finally make the project publicly available: set the Project‘s tag (table: Tag)
- tag "Artenquiz" means it is a quiz
- tag "Public" means it will be publicly accessible
- tag "Identification" make the project available for identification (URL: …/mobile_bestimmung/, TODO: not yet implemented)